Introduction
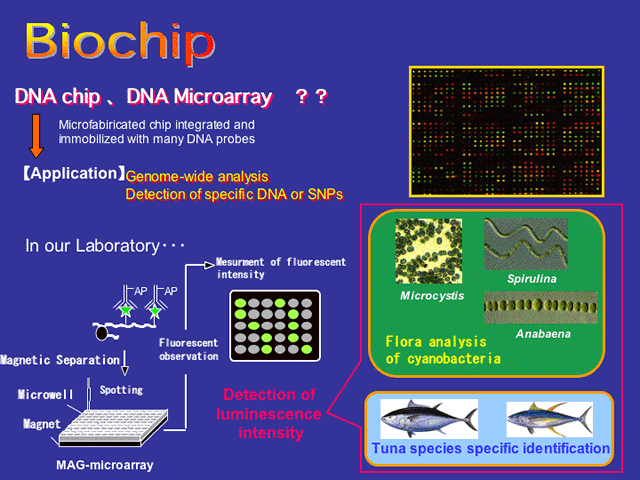
DNA microarray provides rapid, cost-effective, and simultaneous screening of genome DNA for numerous sequence variations. The term DNA microarray generally refers to a gridded array of nucleic acid species on a flat solid support, typically glass or silicon chip devices. Diversified kinds of DNA microarrays have been developed for surveying variations in DNA or RNA and analysis of samples using DNA microarrays is fast becoming a standard approach in molecular biology research and clinical diagnostics. Microarrays have been also used for the investigation of microbial genome in gene expression analysis and quantitation of target microbial populations for environmental analysis.
On the other hand, Micro-Total-Analysis System (m-TAS) and Lab-On-A-Chip technology based on nanotechnology have been recently attracting much attention due to their increasing applications to DNA extraction, DNA amplification and in the screening of newly discovered drugs. These technologies have interesting potential in simple, rapid, cost-effective and highly sensitive analysis for biotechnological processes.
Our group focuses on the research of gSpecies-specific detection using DNA microarray-based techniquesh and gGenome analysis system using On-Chip deviceh.
Discrimination Between Tuna species Using DNA Microarray-Based Techniques
Exact identification of the species and origin of marine products is necessary, particularly for foreign trade. One example would be the northern bluefin tuna (Thunnus thynnus) which is most highly rated as a gsashimih product for the Japanese market. Due to its popularity, illegal fishing and trading have accelerated with the disregard of the decreasing number of the species. However, discrimination of this species from the Pacific subspecies (T. thynnus orientalis) is nearly impossible when the few diagnostic external and internal morphological characteristics are removed, or when they are filleted.
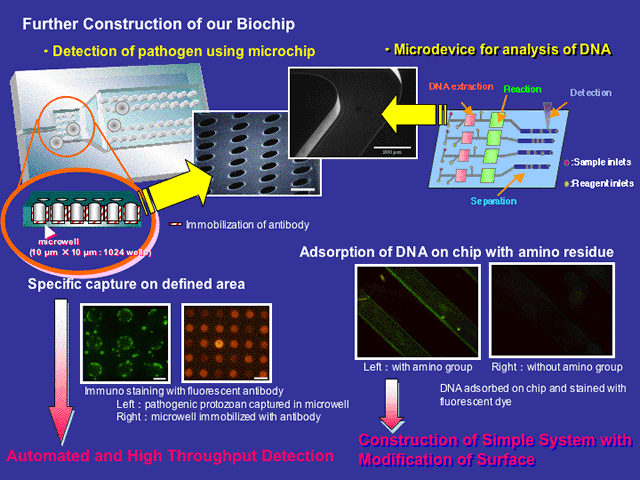
A DNA fragment (ATCO) flanking the mitochondrial ATPase and cytochrome oxidase subunit III wereemployed for the distinction of differences in the restriction profiles and nucleotide sequences between Atlantic and Pacific northern bluefin tunas. The entire nucleotide sequence of the ATCO fragments of these two subspecies were performed in the design of specific DNA probes. A magnetic-capture hybridization technique employing bacterial magnetic particles (BMPs: see Magnetic Bacteria for more detail), which are isolated from the magnetic bacterium, was applied to differentiate the Atlantic and Pacific subspecies of the northern bluefin tuna (Thunnus thynnus) using these specific DNA sequences. The BMPs were concentrated, spotted in 100-Êm-size microwell on microarray, and the signal was detected. This system employing DNA on BMPs may be useful for discrimination of these two subspecies by recognizing a single-nucleotide difference.
DNA Extraction from Whole Blood Based on Electrostatic Interactions
A cascading hyperbranched polyamidoamine dendrimer was synthesized on the surface of BMPs to allow enhanced extraction of DNA from fluid suspensions. Characterization of the synthesis revealed linear doubling of the surface amine charge from generations one through five starting with an amino silane initiator. The dendrimer modified magnetic particles were then used to carry out magnetic separation of DNA. The positively charged dendrimer modified magnetic particles bind DNA through electrostatic interactions resulting in the increase in binding and release efficiencies of the particles with the increase in the number of generations. This extraction method enabled us to purify 33 ng of genomic DNA from 1 mL of blood. We have also developed a fully automated system to fully utilize this technique in high-throughput genomic DNA extraction.
DNA Amplification on Micro-Chip
Subsequent to the development of the DNA extraction method, we proceeded with the construction of a micro-chip for DNA extraction and amplification. An amine-coated micro-chip was constructed for DNA binding assay where the binding capacity of the micro-chip reached to 6 ng of DNA. We then conducted DNA amplification of the alcohol dehydrogenase 2 (ALDH2) gene (176 bp) and amplification of the DNA was successfully identified with a reaction volume of 0.5 mL. The entire process of DNA extraction and DNA amplification will be subsequently performed on the micro-chip in future works.