There is a strong demand for sensors to monitor the environmental state, food safety, or medical condition. We are attempting to develop such biosensors using aptamers or DNA-binding proteins, which recognize specific target molecules. We are engineering biomolecules to provide them with additional useful properties for the development of new biosensors. We study aptamer screening, improvement of aptamer function, and application of aptamers to biosensors. We aim to achieve the commercialization of our technologies.
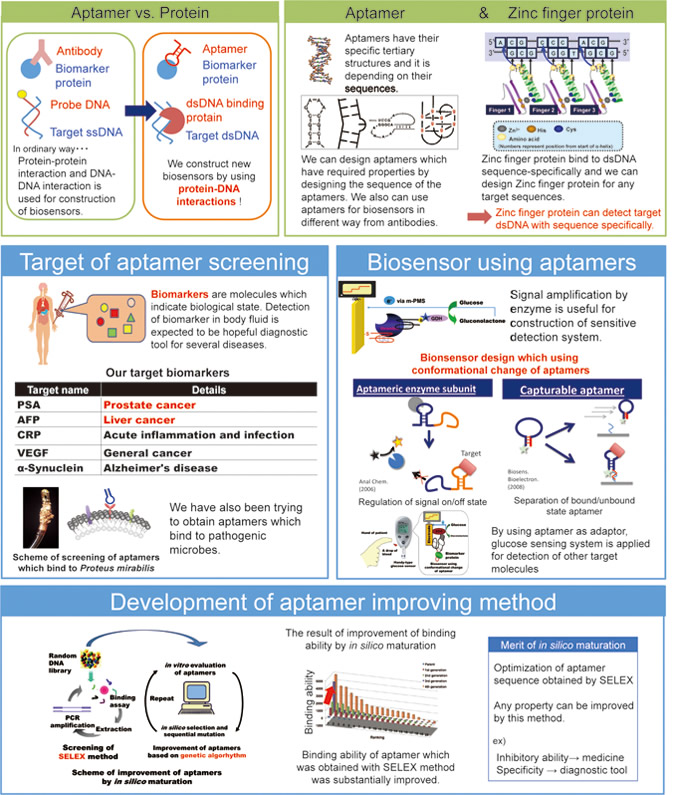

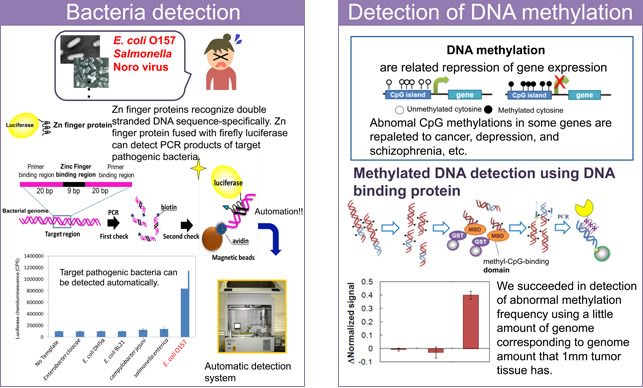
Zinc finger proteins, which are small DNA-binding proteins from mammals, can bind strongly to specific DNA base pairs. We applied this specific DNA recognition of zinc finger proteins in a PCR-based method to detect bacterial genomic DNA. Zinc finger proteins can discriminate specific amplicons from nonspecific amplicons, allowing specific bacterial genome detection.
We succeeded in detection of PCR-amplified DNA fragments from Legionella pneumophila, Escherichia coli O157 and Salmonella species by using zinc finger proteins that recognize a specific sequence within the amplified fragments. Moreover, we have also developed a zinc finger protein-based system for the detection of DNA methylation related to the onset of cancer.

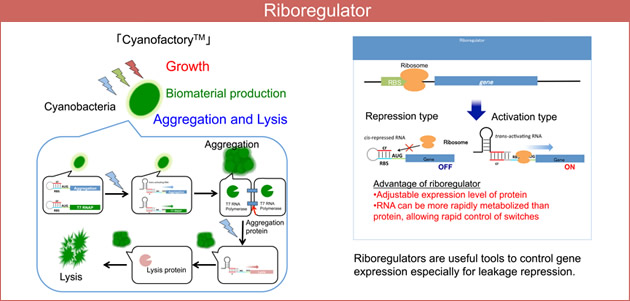
Although microorganisms have long been used for the production of valuable substances, synthetic biological approaches have recently been gathering much attention. Synthetic biology approaches involve the drastic modification of organisms or even the generation of entirely new “synthetic” bacteria. The goal of synthetic biology is generally to create a new material-producing cell by introducing novel metabolic pathways or physiological functions.
Effective control of gene expression is central to achieving these goals. To address this requirement, we focused on RNAs, which can be designed more easily than proteins. Gene expression can be controlled by introducing designed RNA sequences into mRNAs and/or by using RNAs that interact with mRNA. We are constructing functional RNA that works in cyanobacteria and aim to create a new cyanobacteria-based substance-producing system called ‘CyanofactoryTM’.